Accessing a large file using a memory map - numpy memmap (~ 10 times faster)¶
When dealing with large files it could be covenient to avoid loading them entirely into memory but doing it just for the needed segments.
From the documentation: numpy.memmap create a memory-map to an array stored in a binary file on disk, without reading the entire file into memory. NumPy’s memmap’s are array-like objects.
Ghost imaging case
The starting files are 10000 numpy 2D arrays saved in compressed npz files (~470 kB file).
Time needed for a simple task is ~ 15 ms/image (e.g.: accessing all of them, compute a total image, and save the result in a file)
import numpy as np
import matplotlib.pyplot as plt
%%timeit
imtot = np.zeros((700,1000))
for i in range(100):
im = np.load(f'/home/valerio/1slit_thermal_npz/luce_{i:04}.npz')['arr_0'][:700, :1000]
imtot += im
np.save('tot1',imtot)
Writing a big file containing all the needed images (iper-image)
This procedure takes a (relatively) long time once (1.2s for 100 images, final file size 67 MB ):
%%time
import numpy as np
iper = np.zeros((100, 700,1000), dtype='int8')
for i in range(100) :
img = np.load(f'/home/valerio/1slit_thermal_npz/luce_{i:04}.npz')['arr_0'][:700, :1000]
iper[i, :, :] = img
np.save('/home/valerio/Desktop/iper100image', iper)
Reading the "iper-image" file
The time needed for the same task as the previous examples is now:
%%timeit
imtot = np.zeros((100,700,1000))
iperfile = np.load('/home/valerio/Desktop/iper100image.npy','r')
imtot = iperfile.sum(axis=0)
np.save('tot2',imtot)
114 ms against 1.2 s, this method is 10 times faster.
Writing files using memmap (working with bytes)¶
When a big file has to be read, the np.load( ... , mmap_mode='r').
If the memory map mathod has to be used to write files, memmap() has to be used explicitly.
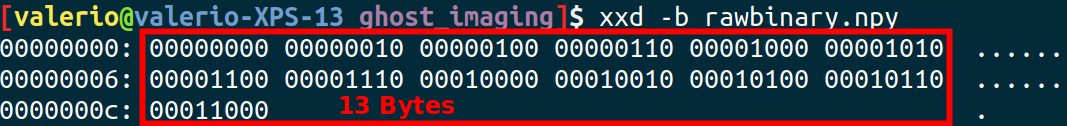
Example creating a (memory map to) file on disk for 10 8-bit integers:
import numpy as np
fp = np.memmap('rawbinary.npy', dtype=np.int8, mode='w+', shape=13)
Write some numbers in the array(-like)
for i in range(13):
fp[i]=i*2
fp
Once done, flush() write any changes in the array to the file on disk.
fp.flush()
Let's check the file size. We expect that 8 bits (or 1 Byte) are used for every integer.
import os
print('The file size is',os.stat('rawbinary.npy').st_size,'bytes' )
Let's check also the content of the binary file (memo: read() import everything in a single string, or bytes with the 'b' option)
file = open('rawbinary.npy', 'rb')
by = file.read()
print('type of by:',type(by))
print('by:',by)
The struct module performs conversions between Python values and C structs represented as Python bytes objects. This can be used in handling binary data stored in files or from network connections, among other sources. It uses Format Strings as compact descriptions of the layout of the C structs and the intended conversion to/from Python values.
import struct
for el in struct.iter_unpack('b',by) :
print(el)
The created file cannot be loaded directly into a numpy array using the load() method, but it is possible to use again a memmap object provided its data type (and shape are known).
This won't work:
a = np.load('rawbinary.npy')
This is ok:
a = np.memmap('rawbinary.npy', dtype=np.int8, mode='r', shape=3)
print(a)
a = np.memmap('rawbinary.npy', dtype=np.int8, mode='r', shape=10)
print(a)
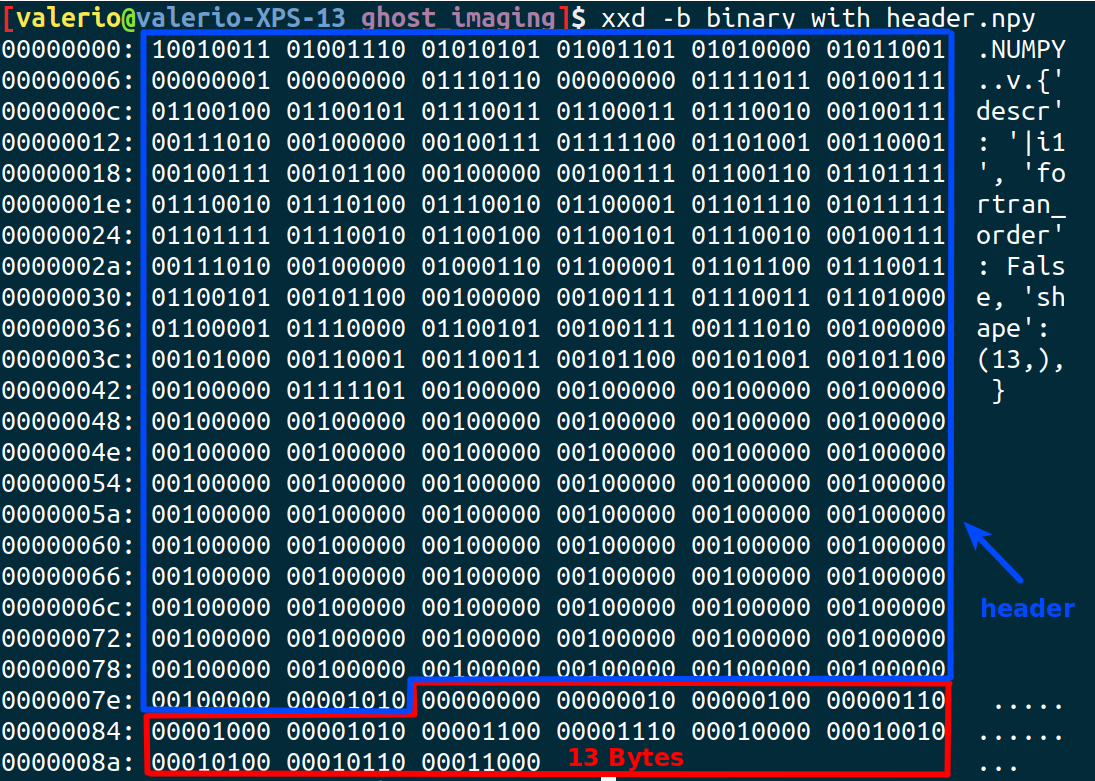
In order to save a file usable with the load() a header must be included to contains also shape and size parameters, that is what save() is doing:
np.save('binary_with_header.npy', fp)
np.load('binary_with_header.npy')